Graphical Abstract
Abstract
Background. Biotin is a water-soluble vitamin that plays a key role in carboxylation. The formation of free biotin is impaired in biotinidase deficiency (BD), resulting in impaired biotin-dependent carboxylase functions. Based on the percentage of residual serum enzyme activity, BD is classified as partial and profound.
Methods. Retrospective data including gender, age, parental consanguinity, family history, biotinidase activity analyses, type of deficiency (partial-profound), physical examination, treatment, and genotypes were evaluated in patients diagnosed with biotinidase deficiency in a single center in the eastern region of Türkiye. Patients whose biotinidase enzyme activity was below 30% with biallelic variants in the BTD gene were diagnosed as BD.
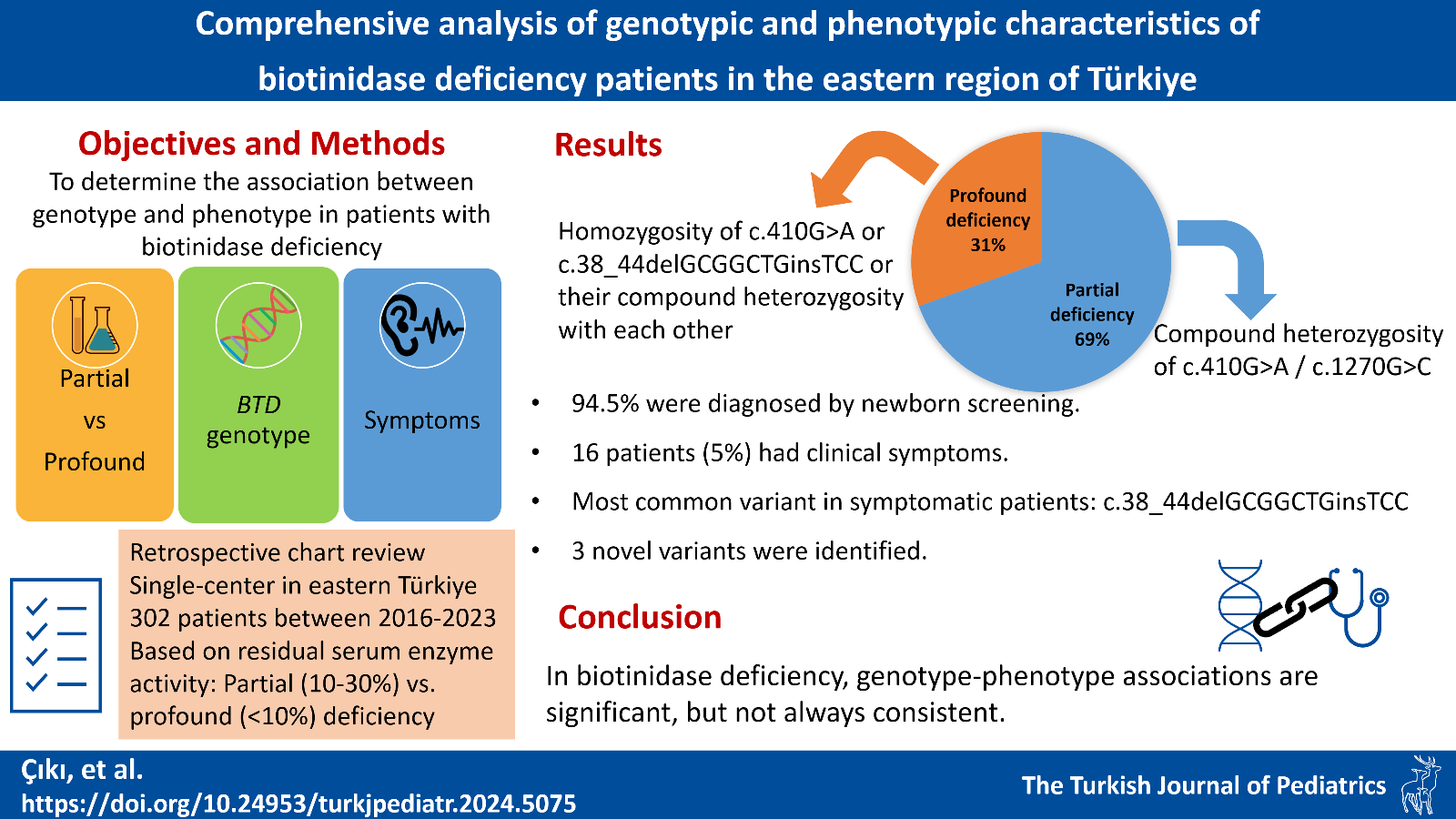
Results. A total of 302 patients were included in the study. Parental consanguinity was present in 135 (44.7%) of them. Two hundred eighty-six (94.7%) were diagnosed by neonatal screening, 14 (4.6%) by family screening and two (0.06%) by clinical symptoms. Ninety-two (30.5%) of the patients were followed-up with profound deficiency and 210 (69.5%) with partial deficiency. A total of 306 variants were detected. Twenty different variants (3 novel - 3 rare) and 31 different genotypes were detected. The 3 most frequently detected variants were c.410G>A (p.Arg137His; 47.3%), c.1270G>C (p.Asp424His; 29.7%), and c.38_44delGCGGCTGinsTCC (p.Cys13Phefs*36; 15.3%). The 3 most frequently identified genotypes were c.410G>A (p.Arg137His) / c.1270G>C (p.Asp424His) compound heterozygous (32.4%), c.410G>A (p.Arg137His) homozygous (24.8%), and c.38_44delGCGGCTGinsTCC (p.Cys13Phefs*36) / c.1270G>C (p.Asp424His) compound heterozygous (12.2%). Patients with c.410G>A (p.Arg137His) homozygous variant, c.38_44delGCGGCTGinsTCC (p.Cys13Phefs*36) homozygous variant and c.38_44delGCGGCTGinsTCC (p.Cys13Phefs*36) / c.410G>A (p.Arg137His) compound heterozygous variant were statistically significantly associated with profound deficiency. Compound heterozygosity of c.410G>A (p.Arg137His) / c.1270G>C (p.Asp424His) variants were significantly associated with partial deficiency.
Conclusions. The association between the BTD genotype and biochemical phenotype is not always consistent. Our study provides valuable data by adding variants with genotype-phenotype correlations to the literature and three novel variants, which can provide significant guidance in clinical follow-up.
Keywords: biotin, biotinidase deficiency, BTD, genotype–phenotype correlation, partial, profound
References
- Canda E, Kalkan Uçar S, Çoker M. Biotinidase deficiency: prevalence, ımpact and management strategies. Pediatric Health Med Ther 2020; 11: 127-133. https://doi.org/10.2147/PHMT.S198656
- Tankeu AT, Van Winckel G, Elmers J, et al. Biotinidase deficiency: what have we learned in forty years? Mol Genet Metab 2023; 138: 107560. https://doi.org/10.1016/j.ymgme.2023.107560
- Wolf B. Biotinidase deficiency. In: Adam MP, Feldman J, Mirzaa GM, et al., editors. GeneReviews®. Seattle, WA: University of Washington; 1993–2024.
- Kannan B, Navamani HK, Jayaseelan VP, Arumugam P. A rare biotinidase deficiency in the pediatrics population: genotype-phenotype analysis. J Pediatr Genet 2022; 12: 1-15. https://doi.org/10.1055/s-0042-1757887
- Tanyalçın T, Aslan D. Use of big data for verification of decision levels for biotinidase deficiency and galactosemia. XXVII. Balkan Clinical Laboratory Federation Meeting BCLF 2019; XXX. National Congress of the Turkish Biochemical Society TBS 2019. Available at: http://www.tanyalcin.com/Data/GALTveBTD_REFERANS_DEGERLER.pdf
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405-424. https://doi.org/10.1038/gim.2015.30
- Wolf B, Grier RE, Allen RJ, Goodman SI, Kien CL. Biotinidase deficiency: the enzymatic defect in late-onset multiple carboxylase deficiency. Clin Chim Acta 1983; 131: 273-281. https://doi.org/10.1016/0009-8981(83)90096-7
- Wolf B, Heard GS, Jefferson LG, Proud VK, Nance WE, Weissbecker KA. Clinical findings in four children with biotinidase deficiency detected through a statewide neonatal screening program. N Engl J Med 1985; 313: 16-19. https://doi.org/10.1056/NEJM198507043130104
- Karaca M, Özgül RK, Ünal Ö, et al. Detection of biotinidase gene mutations in Turkish patients ascertained by newborn and family screening. Eur J Pediatr 2015; 174: 1077-1084. https://doi.org/10.1007/s00431-015-2509-5
- Baykal T, Hüner G, Sarbat G, Demirkol M. Incidence of biotinidase deficiency in Turkish newborns. Acta Paediatr 1998; 87: 1102-1103. https://doi.org/10.1080/080352598750031518
- Toktaş İ, Sarıbaş S, Canpolat S, Erdem Ö, Özbek MN. Evaluation of patients diagnosed with phenylketonuria and biotinidase deficiency by the newborn screening program: a ten-year retrospective study. Turk J Pediatr 2022; 64: 985-992. https://doi.org/10.24953/turkjped.2022.467
- Kasapkara ÇS, Akar M, Özbek MN, et al. Mutations in BTD gene causing biotinidase deficiency: a regional report. J Pediatr Endocrinol Metab 2015; 28: 421-424. https://doi.org/10.1515/jpem-2014-0056
- Yılmaz B, Ceylan AC, Gündüz M, et al. Evaluation of clinical, laboratory, and molecular genetic features of patients with biotinidase deficiency. Eur J Pediatr 2024; 183: 1341-1351. https://doi.org/10.1007/s00431-023-05376-4
- Seker Yilmaz B, Mungan NO, Kor D, et al. Twenty-seven mutations with three novel pathologenic variants causing biotinidase deficiency: a report of 203 patients from the southeastern part of Turkey. J Pediatr Endocrinol Metab 2018; 31: 339-343. https://doi.org/10.1515/jpem-2017-0406
- Canda E, Yazici H, Er E, et al. Single center experience of biotinidase deficiency: 259 patients and six novel mutations. J Pediatr Endocrinol Metab 2018; 31: 917-926. https://doi.org/10.1515/jpem-2018-0148
- Erdol S, Kocak TA, Bilgin H. Evaluation of 700 patients referred with a preliminary diagnosis of biotinidase deficiency by the national newborn metabolic screening program: a single-center experience. J Pediatr Endocrinol Metab 2023; 36: 555-560. https://doi.org/10.1515/jpem-2023-0003
- Oz O, Karaca M, Atas N, Gonel A, Ercan M. BTD gene mutations in biotinidase deficiency: genotype-phenotype correlation. J Coll Physicians Surg Pak 2021; 31: 780-785. https://doi.org/10.29271/jcpsp.2021.07.780
- Akgun A, Sen A, Onal H. Clinical, biochemical and genotypical characteristics in biotinidase deficiency. J Pediatr Endocrinol Metab 2021; 34: 1425-1433. https://doi.org/10.1515/jpem-2021-0242
- Sürücü Kara İ, Köse E, Koç Yekedüz M, Eminoğlu FT. A different approach to the evaluation of the genotype-phenotype relationship in biotinidase deficiency: repeated measurement of biotinidase enzyme activity. J Pediatr Endocrinol Metab 2023; 36: 1061-1071. https://doi.org/10.1515/jpem-2023-0337
- Sharma R, Kucera CR, Nery CR, Lacbawan FL, Salazar D, Tanpaiboon P. Biotinidase biochemical and molecular analyses: experience at a large reference laboratory. Pediatr Int 2024; 66: e15726. https://doi.org/10.1111/ped.15726
- Borsatto T, Sperb-Ludwig F, Lima SE, et al. Biotinidase deficiency: genotype-biochemical phenotype association in Brazilian patients. PLoS One 2017; 12: e0177503. https://doi.org/10.1371/journal.pone.0177503
- Forny P, Wicht A, Rüfenacht V, Cremonesi A, Häberle J. Recovery of enzyme activity in biotinidase deficient individuals during early childhood. J Inherit Metab Dis 2022; 45: 605-620. https://doi.org/10.1002/jimd.12490
- Swango KL, Demirkol M, Hüner G, et al. Partial biotinidase deficiency is usually due to the D444H mutation in the biotinidase gene. Hum Genet 1998; 102: 571-575. https://doi.org/10.1007/s004390050742
- Tanyalcin I, Stouffs K, Daneels D, et al. Convert your favorite protein modeling program into a mutation predictor: “MODICT”. BMC Bioinformatics 2016; 17: 425. https://doi.org/10.1186/s12859-016-1286-0
- Hsu RH, Chien YH, Hwu WL, et al. Genotypic and phenotypic correlations of biotinidase deficiency in the Chinese population. Orphanet J Rare Dis 2019; 14: 6. https://doi.org/10.1186/s13023-018-0992-2
- Saleem H, Simpson B. Biotinidase deficiency. In: StatPearls. Treasure Island, FL: StatPearls Publishing; 2024. Available at: https://www.ncbi.nlm.nih.gov/books/NBK560607/
- Wolf B. Biotinidase deficiency: “if you have to have an inherited metabolic disease, this is the one to have”. Genet Med 2012; 14: 565-575. https://doi.org/10.1038/gim.2011.6
Copyright and license
Copyright © 2024 The Author(s). This is an open access article distributed under the Creative Commons Attribution License (CC BY), which permits unrestricted use, distribution, and reproduction in any medium or format, provided the original work is properly cited.















